- 2025
- 2024
- 2023
- 2022
- 2021
- 2020
- 2019
- Pre-2019
Downie, L., Ferrandiz, N., Courthold, E., Jones, M., and Royle, S.J. (2025)
Nondisruptive inducible labeling of ER-membrane contact sites using the Lamin B receptor
PLoS Biol 23, e3003249
[Link]
Scott, W., Polutranko, V., Milczarek, J., Hands-Portman, I., and Balasubramanian, M.K. (2025)
Fluorescent protein tags for human tropomyosin isoform comparison
Biol Open bio.061992
[Link]
Morales Angeles, D., Coleman, K., Progress Odika, C., L B Graham, C., Chan, H., Gilmore, M., et al. (2025)
SpoIIIL is a forespore factor required for efficient cell-cell signalling during Bacillus subtilis sporulation
PLoS Genet 21, e1011768
[Link]
Mohanrao, R., Pinto, C.S., Suchenko, A., Clarkson, G.J., Wills, M., Roesner, S., et al. (2025)
Single-Benzene-Based Clickable Fluorophores for In Vitro and In Vivo Bioimaging
ChemistrySelect 10, slct.202405738
[Link]
Inman, A., Spiritosanto, E., Evans, B.L., Lutton, J.E., Tada, M., Bretschneider, T., et al. (2025)
A multi-tiered mechanical mechanism shapes the early neural plate
Nat Commun 16, 6187
[Link]
Fesenko, M., and Royle, S.J. (2025)
Small but mighty: ATG9A-positive vesicles are a branch of the intracellular nanovesicle superfamily
Autophagy Rep 4, 2513467
[Link]
Fesenko, M., Moore, D.J., Ewbank, P., Courthold, E., and Royle, S.J. (2025)
ATG9A vesicles are a subtype of intracellular nanovesicle
J Cell Sci 138, jcs263852
[Link]
Abid Ali, F., Zwetsloot, A.J., Stone, C.E., Morgan, T.E., Wademan, R.F., Carter, A.P., and Straube, A. (2025)
KIF1C activates and extends dynein movement through the FHF cargo adapter
Nat Struct Mol Biol 32, 756–766
[Link]
Chew, Y.-M., and Cross, R.A. (2025)
Structural switching of tubulin in the microtubule lattice
Biochem Soc Trans 53, BST20240360
[Link]
Wohland, T., Saunders, T.E., and Chan, C.J. (2025)
Developmental biophysics
Biophys J 124, E1–E2
[Link]
MacDonald, E., Forrester, A., Valades-Cruz, C.A., Madsen, T.D., Hetmanski, J.H.R., Dransart, E., Ng, Y., Godbole, R., Shp, A.A., Leconte, L., et al. (2025)
Growth factor-triggered de-sialylation controls glycolipid-lectin-driven endocytosis
Nat Cell Biol 27, 449–463
[Link]
Shelford, J., Burgess, S.G., Rostkova, E., Richards, M.W., Larocque, G., Sampson, J., Tiede, C., Fielding, A.J., Daviter, T., Tomlinson, D.C., et al. (2025)
Structural characterization and inhibition of the interaction between ch-TOG and TACC3
J Cell Biol 224, e202407002
[Link]
Souza, D.P., Espadas, J., Chaaban, S., Moody, E.R.R., Hatano, T., Balasubramanian, M., Williams, T.A., Roux, A., and Baum, B. (2025)
Asgard archaea reveal the conserved principles of ESCRT-III membrane remodeling
Sci Adv 11, eads5255
[Link]
R Mohanrao, CS Pinto, A Suchenko, GJ Clarkson, M Wills, S Roesner, ... (2025)
Single‐Benzene‐Based Clickable Fluorophores for In Vitro and In Vivo Bioimaging
ChemistrySelect 10 (9), e202405738
[Link]
A Paterou, J Sáez Conde, J Týč, JD Sunter, S Vaughan, K Gull, S Dean (2025)
A comprehensive toolkit for protein localization and functional analysis in trypanosomatids
Open Biology 15 (4), 240361
[Link]
C Spahn, S Middlemiss, E Gómez-de-Mariscal, R Henriques, HB Bode, ... (2025)
The nucleoid of rapidly growing Escherichia coli localizes close to the inner membrane and is organized by transcription, translation, and cell geometry
Nature Communications 16 (1), 3732
[Link]
SC Al-Izzi, SG Nodehi, DV Köster, RG Morris (2025)
ATP-controlled remodeling in reconstituted actomyosin
Physical Review Research 7 (1), 013175
[Link]
SJ Bray, SJ Royle, HA Shiels, D St Johnston (2025)
The Company of Biologists: celebrating 100 years
Biol Open 14(1):BIO061842.
[Link]
S Zhu, YT Loo, S Veerapathiran, TYJ Loo, BN Tran, C Teh, J Zhong, ... (2025)
Receptor binding and tortuosity explain morphogen local-to-global diffusion coefficient transition
Biophysical Journal 124 (6), 963-979
[Link]
S Tlili, M Shagirov, S Zhang, TE Saunders (2025)
Interfacial energy constraints are sufficient to align cells over large distances
Biophysical Journal 124 (6), 1011-1023
[Link]
TE Saunders (2025)
Topology drives cell fate
Nature Physics volume 21, pages 508–509
[Link]
Y Le, K Rajasekhar, TYJ Loo, TE Saunders, T Wohland, C Winkler (2025)
Midkine-a interacts with Ptprz1b to regulate neural plate convergence and midline formation in the developing zebrafish hindbrain
Developmental Biology 521:52-74. doi: 10.1016/j.ydbio.2025.02.004
[Link]
de-Carvalho, J., Tlili, S., Saunders, T.E., and Telley, I.A. (2024)
The positioning mechanics of microtubule asters in Drosophila embryo explants
Elife 12, RP90541
[Link]
Huang, H.-L., Suchenko, A., Grandinetti, G., Balasubramanian, M.K., Chinthalapudi, K., and Heissler, S.M. (2024)
Cryo-EM structures of cardiac muscle α-actin mutants M305L and A331P give insights into the structural mechanisms of hypertrophic cardiomyopathy
Eur J Cell Biol 103, 151460
[Link]
Ivorra-Molla, E., Akhuli, D., McAndrew, M.B.L., Scott, W., Kumar, L., Palani, S., Mishima, M., Crow, A., and Balasubramanian, M.K. (2024)
A monomeric StayGold fluorescent protein
Nat Biotechnol 42, 1368–1371
[Link]
Ngo, K.X., Vu, H.T., Umeda, K., Trinh, M.-N., Kodera, N., and Uyeda, T. (2024)
Deciphering the actin structure-dependent preferential cooperative binding of cofilin
Elife 13, RP95257
[Link]
Straube, A., and Tanner, K. (2024)
Machines, mechanics and mechanisms of cells and tissues
Curr Opin Cell Biol 88, 102346
[Link]
Sumiyoshi, R., Yamagishi, M., Furuta, A., Nishizaka, T., Furuta, K., Cross, R.A., and Yajima, J. (2024)
Tether-scanning the kinesin motor domain reveals a core mechanical action
Proc Natl Acad Sci U S A 121, e2403739121
[Link]
Avissar-Whiting, M., Belliard, F., Bertozzi, S. M., Brand, A., Brown, K., Clément-Stoneham, G., Dawson, S., Dey, G., Ecer, D., Edmunds, S. C., Farley, A., Fischer, T. D., Franko, M., Fraser, J. S., Funk, K., Ganier, C., Harrison, M., Hatch, A., Hazlett, H., Hindle, S.,
Hook, D.W., Hurst, P., Kamoun, S., Kiley, R., Lacy, M.M., LaFlamme, M., Lawrence, R., Lemberger, T., Leptin, M., Lumb, E., MacCallum, C.J., Marcum, C.S., Marinello, G., Mendonça, A., Monaco, S., Neves, K., Pattinson, D., Polka, J.K., Puebla, I., Rittman, M.,
Royle, S.J., Saderi, D., Sever, R., Shearer, K., Spiro, J.E., Stern, B., Taraborelli, D., Vale, R., Vasquez, C.G., Waltman, L., Watt, F.M., Weinberg, Z.Y. & Williams, M. (2024)
Recommendations for accelerating open preprint peer review to improve the culture of science.
PLoS Biology, 22(2), e3002502.
[Link]
Mihalas BP, Pieper GH, Aboelenain M, Munro L, Srsen V, Currie CE, Kelly DA, Hartshorne GM, Telfer EE, McAinsh AD, Anderson RA, Marston AL.
Age-dependent loss of cohesion protection in human oocytes.
Curr Biol. 2024 Jan 8;34(1):117-131.e5.
[Link]
Sittewelle, M. & Royle, S.J. (2024)
Passive diffusion accounts for the majority of intracellular nanovesicle transport
Life Sci Alliance 7 (1) e202302406
[Link]
------------------
S Forshaw, JS Parker, WT Scott, RC Knighton, N Tiwari, SM Oladeji, ...(2024)
Increasing the versatility of the biphenyl-fused-dioxacyclodecyne class of strained alkynes
Organic & Biomolecular Chemistry 22 (3), 590-605
Tzamarias, Byron, Ballesta, Annabelle, Burroughs, Nigel John (2024)
Aperiodic optimal chronotherapy in simple compartment tumour growth models under circadian drug toxicity conditions
Journal Article 11 November 2024 Mathematics MDPI 12 22 10.3390/math12223516
https://doi.org/10.3390/math12223516 2227-7390 189303 16 Jan 2025 14:37 16 Jan 2025 14:37 Public Open Access (Creative Commons open licence) 6 November 2024 11 November 2024 15 January 2025 16 January 2025
Burroughs, Nigel J., Leuridan, Mathilde L. C. (2024)
Optimising the tumour elimination payoff in cancer therapy
Journal Article 23 September 2024 IET Control Theory & Applications The Institution of Engineering and Technology 18 13 pp. 1621-1637 10.1049/cth2.12701 https://doi.org/10.1049/cth2.12701 1751-8644 189304 16 Jan 2025 14:53 16 Jan 2025 14:53 Public Open Access (Creative Commons open licence) 28 May 2024 23 June 2024 15 January 2025 16 January 2025
H Mamar, R Fajka-Boja, M Mórocz, EP Jurado, S Zentout, A Mihuţ, ... (2024)
The loss of DNA polymerase epsilon accessory subunits POLE3–POLE4 leads to BRCA1-independent PARP inhibitor sensitivity
Nucleic Acids Research 52 (12), 6994-7011
RR Kay, JE Lutton, JS King, T Bretschneider (2024)
Making cups and rings: the ‘stalled-wave’model for macropinocytosis
Biochemical Society Transactions 52 (4), 1785-1794
E Offord, J Lutton, T Bretschneider (2024)
Heterogeneous Graph Neural Networks for Analysing Spatio-Temporal Cell Surface Dynamics
2024 IEEE International Symposium on Biomedical Imaging (ISBI), 1-5
CelFDrive: Artificial Intelligence assisted microscopy for automated detection of rare events 1 2024
A Paterou, JS Conde, S Neupane, S Dean (2024)
The ABCD_AF291 antibody gives strong and specific signal in ultra-expansion microscopy analysis of HA tagged proteins in African trypanosomes
Antibody Reports 7 (1), e1625-e1625
G Benn, C Bortolini, DM Roberts, ALB Pyne, S Holden, BW Hoogenboom (2024)
Complement-mediated killing of Escherichia coli by mechanical destabilization of the cell envelope
The EMBO Journal 43 (23), 6152-6160
C Vanhille-Campos, KD Whitley, P Radler, M Loose, S Holden, A Šarić (2024)
Self-organization of mortal filaments and its role in bacterial division ring formation
Nature Physics 20 (10), 1670-1678
S Middlemiss, M Blandenet, DM Roberts, A McMahon, J Grimshaw, ... (2024)
Molecular motor tug-of-war regulates elongasome cell wall synthesis dynamics in Bacillus subtilis
Nature Communications 15 (1), 5411
KD Whitley, J Grimshaw, DM Roberts, E Karinou, PJ Stansfeld, S Holden (2024)
Peptidoglycan synthesis drives a single population of septal cell wall synthases during division in Bacillus subtilis
Nature Microbiology 9 (4), 1064-1074
F Torta, N Hoffmann, B Burla, I Alecu, M Arita, T Bamba, SAL Bennett, ... (2024)
Concordant inter-laboratory derived concentrations of ceramides in human plasma reference materials via authentic standards
Nature communications 15 (1), 8562
MS Asokan, RF Joan, S Babji, G Dayma, P Nadukkandy, ... (2024)
Immunogenicity of SARS-CoV-2 vaccines BBV152 (COVAXIN®) and ChAdOx1 nCoV-19 (COVISHIELD™) in seronegative and seropositive individuals in India: a multicentre, nonrandomised …
The Lancet Regional Health-Southeast Asia 22
MF Garcia-Parajo, S Mayor (2024)
The ubiquitous nanocluster: A molecular scale organizing principle that governs cellular information flow
Current opinion in cell biology 86, 102285
JA Theriot, A Simonsen, I Tolić, MD Leonetti, S Mayor, P Bassereau, ... (2024)
Cell biology is…
Cell 187 (2), 219-224
A Daniyan, AV Inchingolo, A McAinsh, N Burroughs (2024)
Enhanced Kinetochore Detection During Mitotic Human Cell Division using CFAR
2024 27th International Conference on Information Fusion (FUSION), 1-7
O-070
A Byrska, C Currie, D Taylor, M Erent, A Inchingolo, G Hartshorne, ... (2024)
Cas9-based chromosome labelling reveals large-scale age-dependent centromere reorganisation in human oocytes
Human Reproduction 39 (Supplement_1), deae108. 076
GI Mashanov, JE Molloy (2024)
Single molecule dynamics in a virtual cell combining a 3-dimensional matrix model with random walks
Scientific Reports 14 (1), 20032
CT Chien, H Puhl, SS Vogel, JE Molloy, W Chiu, S Khan (2024)
Hub stability in the calcium calmodulin-dependent protein kinase II
Communications Biology 7 (1), 766
S Khan, JE Molloy, H Puhl, H Schulman, SS Vogel (2024)
Real-time single-molecule imaging of CaMKII-calmodulin interactions
Biophysical Journal 123 (7), 824-838
TE Saunders, RA Cross, AJ Bowman (2024)
Designing and Delivering an Interdisciplinary Undergraduate Degree in Quantitative Biology
The Biophysicist
J de-Carvalho, S Tlili, TE Saunders, IA Telley (2024)
The positioning mechanics of microtubule asters in Drosophila embryo explants
Elife 12, RP90541
T Athilingam, AVS Nelanuthala, C Breen, N Karedla, M Fritzsche, ... (2024)
Long-range formation of the Bicoid gradient requires multiple dynamic modes that spatially vary across the embryo
Development 151 (3)
TJ Wherley, S Thomas, DP Millay, T Saunders, S Roy (2024)
Molecular regulation of myocyte fusion
Current topics in developmental biology 158, 53-82
Hill, Edward M., Ryan, Matthew, and Haw, David et al.(2024)
Integrating human behaviour and epidemiological modelling : unlocking the remaining challenges
Journal Article 2024 Mathematics in Medical and Life Sciences Routledge 1 1 10.1080/29937574.2024.2429479 https://doi.org/10.1080/29937574.2024.2429479 2993-7574 188454 08 Nov 2024 15:11 04 Mar 2025 14:28 Public Open Access (Creative Commons open licence) 4 November 2024 29 November 2024 8 November 2024 27 February 2025
Nagorska A, Zaucker A, Lambert F, Inman A, Toral-Perez S, Gorodkin J, Wan Y, Smutny M, Sampath K.
Translational control of furina by an RNA regulon is important for left-right patterning, heart morphogenesis and cardiac valve function.
Development. 2023 Dec 1;150(23):dev201657. doi: 10.1242/dev.201657.
[Link]
Ivorra-Molla E, Akhuli D, McAndrew MBL, Scott W, Kumar L, Palani S, Mishima M, Crow A, Balasubramanian MK.
A monomeric StayGold fluorescent protein.
Nat Biotechnol. 2023 Dec 11. doi: 10.1038/s41587-023-02018-w. Online ahead of print.
Hamshaw, I., Straube, A., Stark, R., Baxter, L., Alam, M.T., Wever, W.J., Yin, J., Yue, Y., Pinton, P., Sen, A., Feguson, G.D. and Blanks, A.M. (2023)
PGF2α induces a pro-labour phenotypical switch in human myometrial cells that can be inhibited with PGF2α receptor antagonists.
Front. Pharmacol. 14:1285779.
[Link]
Mosby, L.S., Straube, A. & Polin, M. (2023)
A general model for the motion of multivalent cargo interacting with substrates.
J. R. Soc. Interface 20: 20230510.
[Link]
Chew Y-M & Cross R.A. (2023)
Taxol acts differently on different tubulin isotypes
Comms. Biol. (6) 946
[Link]
Sittewelle, M., Ferrandiz, N., Fesenko, M. & Royle, S.J. (2023)
Genetically encoded imaging tools for investigating cell dynamics at a glance
J. Cell Sci. 136 (7): jcs260783
[Link]
McAinsh AD, Kops GJPL. (2023)
Principles and dynamics of spindle assembly checkpoint signalling
Nat Rev Mol Cell Biol. Mar 24. doi: 10.1038/s41580-023-00593-z.
[Link]
Straube A. (2023)
The cell biology of motors
J. Cell Sci. 136(5): jcs261056. doi: 10.1242/jcs.261056.
[Link]
Arora AS, Huang HL, Singh R, Narui Y, Suchenko A, Hatano T, Heissler SM, Balasubramanian MK, Chinthalapudi K. (2023)
Structural insights into actin isoforms.
Elife. 12:e82015. doi: 10.7554/eLife.82015.
[Link]
E Ivorra-Molla, D Akhuli, MBL McAndrew, W Scott, L Kumar, S Palani, ... (2023)
A monomeric StayGold fluorescent protein
Nature Biotechnology, 1-4
[Link]
Peter Nietmann, Kevin Kaub, Andrejus Suchenko, Susanne Stenz, Claas Warnecke, Mohan K. Balasubramanian & Andreas Janshoff (2023)
Cytosolic actin isoforms form networks with different rheological properties that indicate specific biological function
Nature Communications 14 (1), 7989
[Link]
Amandeep S Arora, Hsiang-Ling Huang, Ramanpreet Singh, Yoshie Narui, Andrejus Suchenko, Tomoyuki Hatano, Sarah M Heissler,Mohan K Balasubramanian & Krishna Chinthalapudi (2023)
Structural insights into actin isoforms
Elife 12, e82015
[Link]
J.E. Lutton, H.L.E. Coker, P. Paschke, C.J. Munn, J.S. King, T. Bretschneider & R.R. Kay (2023)
Formation and closure of macropinocytic cups in Dictyostelium
Current Biology 33 (15), 3083-3096. e6
[Link]
E Offord, EJ Lutton, T Bretschneider (2023)
Cell membrane feature detection using graph neural networks
IEEE xplore
[Link]
A Platt, EJ Lutton, E Offord, T Bretschneider (2023)
MiCellAnnGELo: Annotate microscopy time series of complex cell surfaces with 3D Virtual Reality
Bioinformatics 39 (1), btad013
[Link]
N Siddiqui, D Roth, A Toleikis, AJ Zwetsloot, RA Cross, A Straube (2023)
Force generation of KIF1C is impaired by pathogenic mutations
Current Biology 32 (17), 3862-3870. e6 2023
[Link]
J. Pyrih, M. Hammond, A. Alves, S. Dean, J. Daniel, S. Richard, J. Wheeler, K. Gull & J. Luke (2023)
Comprehensive sub-mitochondrial protein map of the parasitic protist Trypanosoma brucei defines critical features of organellar biology
Cell reports 42 (9)
[Link]
JD Sunter, S Dean, RJ Wheeler (2023)
TrypTag. org: from images to discoveries using genome-wide protein localisation in Trypanosoma brucei
Trends in Parasitology
[Link]
K Billington, C Halliday, R Madden, P Dyer, AR Barker, FF Moreira-Leite, ... (2023)
Genome-wide subcellular protein map for the flagellate parasite Trypanosoma brucei
Nature microbiology 8 (3), 533-547
[Link]
C Halliday, S Dean, JD Sunter, RJ Wheeler (2023)
Subcellular protein localisation of Trypanosoma brucei bloodstream form-upregulated proteins maps stage-specific adaptations
Wellcome Open Research 8
[Link]
Whitley KD, Grimshaw J, Holden S. (2023)
Watching Bacterial Cell Division One Molecule at a Time in Vertical Cells
Microsc Microanal. 2023 Jul 22;29(29 Suppl 1):1070. doi: 10.1093/micmic/ozad067.549. PMID: 37613179
[Link]
Ivorra-Molla E, Akhuli D, McAndrew MBL, Scott W, Kumar L, Palani S, Mishima M, Crow A, Balasubramanian MK. (2023)
A monomeric StayGold fluorescent protein
Nat Biotechnol. 2023 Dec 11. doi: 10.1038/s41587-023-02018-w. PMID: 38081970
[Link]
McAinsh AD, Kops GJPL. (2023)
Principles and dynamics of spindle assembly checkpoint signalling
Nat Rev Mol Cell Biol. Mar 24. doi: 10.1038/s41580-023-00593-z.
[Link]
NAW Bell, JE Molloy (2023)
Single-molecule force spectroscopy reveals binding and bridging dynamics of PARP1 and PARP2 at DNA double-strand breaks
Proceedings of the National Academy of Sciences 120 (22), e2214209120
[Link]
A Meli, A McCormack, I Conte, Q Chen, J Streetley, ML Rose, R Bierings, ...(2023)
Altered storage and function of von Willebrand factor in human cardiac microvascular endothelial cells isolated from recipient transplant hearts
International Journal of Molecular Sciences 24 (5), 4553 2023
[Link]
L Mosby, A Straube, M Polin (2023)
A general model for the motion of multivalent cargo interacting with substrates
Journal of the Royal Society Interface 20 (208), 20230510 2023
[Link]
NAM Araújo, LMC Janssen, T Barois, G Boffetta, I Cohen, A Corbetta, ... (2023)
Steering self-organisation through confinement
Soft matter 19 (9), 1695-1704 2023
[Link]
N Ferrandiz, SJ Royle (2023)
3D Ultrastructural Visualization of Mitosis Fidelity in Human Cells Using Serial Block Face Scanning Electron Microscopy (SBF-SEM)
Bio-protocol 13 (13) 2023
[Link]
M Sittewelle, N Ferrandiz, M Fesenko, SJ Royle (2023)
Genetically encoded imaging tools for investigating cell dynamics at a glance
Journal of Cell Science 136 (7), jcs260783 2023
[Link]
Nagorska A, Zaucker A, Lambert F, Inman A, Toral-Perez S, Gorodkin J, Wan Y, Smutny M, Sampath K. (2023)
Translational control of furina by an RNA regulon is important for left-right patterning, heart morphogenesis and cardiac valve function.
Development. 2023 Dec 1;150(23):dev201657. doi: 10.1242/dev.201657. PMID: 38032088
[Link]
PJY Toh, M Sudol, TE Saunders (2023)
Optogenetic control of YAP can enhance the rate of wound healing
Cellular & Molecular Biology Letters 28 (1), 39 3
[Link]
Y Lou, JF Rupprecht, S Theis, T Hiraiwa, TE Saunders (2023)
Curvature-induced cell rearrangements in biological tissues
Physical Review Letters 130 (10), 108401 5
[Link]
K Karkali, TE Saunders, G Panayotou, E Martín-Blanco (2023)
JNK signaling in pioneer neurons organizes ventral nerve cord architecture in Drosophila embryos
Nature Communications 14 (1), 675 7
[Link]
A Nagorska, A Zaucker, F Lambert, A Inman, S Toral-Perez, J Gorodkin, ... (2023)
Translational control of furina by an RNA regulon is important for left-right patterning, heart morphogenesis and cardiac valve function
Development 150 (23)
[Link]
LS Mosby, A Straube, M Polin (2023)
A general model for the motion of multivalent cargo interacting with substrates
Journal of the Royal Society Interface 20 (208), 20230510 2023
[Link]
A Straube (2023)
Microtubules and microtubule associated proteins (MAPs)
Encyclopedia of cell biology Second edition Academic Press 3, 6-16 4
[Link]
A Straube, FA Ali, A Zwetsloot, C Stone, T Morgan, R Wademan, A Carter (2023)
KIF1C activates and extends dynein movement through the FHF cargo adaptor
[Link]
M Taher, A Straube, A Noel (2023)
Generalized Model of Neurite Trafficking
Proceedings of the 10th ACM International Conference on Nanoscale Computing …
[Link]
A Straube (2023)
The cell biology of motors
Journal of Cell Science 136 (5), jcs261056
[Link]
A Straube (2023)
Interview with the Guest Editor–Anne Straube
Journal of Cell Science 136 (5)
[Link]
SK Schnyder, JJ Molina, R Yamamoto, MS Turner (2023)
Rational social distancing policy during epidemics with limited healthcare capacity
PLOS Computational Biology 19 (10), e1011533
[Link]
K Sankaewtong, JJ Molina, MS Turner, R Yamamoto (2023)
Learning to swim efficiently in a nonuniform flow field
Physical Review E 107 (6), 065102
[Link]
JO Law, CM Jones, T Stevenson, TA Williamson, MS Turner, ...(2023)
A bending rigidity parameter for stress granule condensates
Science Advances 9 (20), eadg0432 1
[Link]
C Feng, JJ Molina, MS Turner, R Yamamoto (2023)
Dynamics of microswimmers near a liquid–liquid interface with viscosity difference
Physics of Fluids 35 (5) 3
[Link]
HL Devereux, MS Turner (2023)
Environmental path-entropy and collective motion
Physical Review Letters 130 (16), 168201 1
[Link]
S Imamura, K Sawaki, JJ Molina, MS Turner, R Yamamoto (2023)
Collective Motion of Quincke Rollers with Fully Resolved Hydrodynamics
Advanced Theory and Simulations, 2200683 2
[Link]
Pardal A.J. & Bowman A.J. (2022)
A specific role for importin-5 and NASP in the import and nuclear hand-off of monomeric H3.
eLife 11:e81755 [Link]
Embacher, P.A., Germanova, T.E., Roscioli, E., McAinsh, A.D. & Burroughs, N.J. (2022)
Bayesian inference of multi-point macromolecular architecture mixtures at nanometre resolution.
PLoS Comput Biol. 18(12):e1010765 [Link]
Sugawa, M., Maruyama, Y., Yamagishi, M., Cross, R.A. & Yajima, J. (2022)
Motor generated torque drives coupled yawing and orbital rotations of kinesin coated gold nanorods
Commun Biol. 5(1):1368 [Link]
Currie, C.E., Ford, E., Benham Whyte, L., Taylor, D.M., Mihalas, B.P., Erent, M., Marston, A.L., Hartshorne, G.M. & McAinsh, A.D. (2022)
The first mitotic division of human embryos is highly error prone
Nat Commun. 13(1):6755 [Link]
Chin, S.M., Hatano, T., Sivashanmugam, L., Suchenko, A., Kashina, A.S., Balasubramanian, M.K. & Jansen, S. (2022)
N-terminal acetylation and arginylation of actin determines the architecture and assembly rate of linear and branched actin networks.
J Biol Chem. 298(11):102518 [Link]
Hatano, T., Lim, T.C., Billault-Chaumartin, I., Dhar, A., Gu, Y., Massam-Wu, T., Scott, W., Adishesha, S., Chapa-Y-Lazo, B., Springall, L., Sivashanmugam, L., Mishima, M., Martin, S.G., Oliferenko, S., Palani, S. & Balasubramanian, M.K. (2022)
mNG-tagged fusion proteins and nanobodies to visualize tropomyosins in yeast and mammalian cells.
J Cell Sci. 135(18):jcs260288 [Link]
McAinsh, A.D. & Marston, A.L. (2022)
The Four Causes: The Functional Architecture of Centromeres and Kinetochores
Annu Rev Genet. doi: 10.1146/annurev-genet-072820-034559.
[Link]
Castrogiovanni, C., Inchingolo, A.V., Harrison, J.U., Dudka, D., Sen, O., Burroughs, N.J., McAinsh, A.D. & Meraldi, P. (2022)
Evidence for a HURP/EB free mixed-nucleotide zone in kinetochore-microtubules
Nat Commun. 13(1):4704. doi: 10.1038/s41467-022-32421-x
[Link]
Siddiqui, N., Roth, D., Toleikis, A., Zwetsloot, A.J., Cross, R.A. & Straube, A. (2022)
Force generation of KIF1C is impaired by pathogenic mutations
Current Biology 32 [Link]
Küey, C., Sittewelle, M., Larocque, G., Hernández-González, M. & Royle, S.J. (2022)
Recruitment of clathrin to intracellular membranes is sufficient for vesicle formation
eLife, doi: 10.7554/eLife.78929
[Link]
Toh PJY, Lai JKH, Hermann A, Destaing O, Sheetz MP, Sudol M, Saunders TE. (2022)
Optogenetic control of YAP cellular localisation and function
EMBO Rep. 23(9):e54401. doi: 10.15252/embr.202154401.
[Link]
Mendieta-Serrano MA, Dhar S, Ng BH, Narayanan R, Lee JJY, Ong HT, Toh PJY, Röllin A, Roy S, Saunders TE. (2022)
Slow muscles guide fast myocyte fusion to ensure robust myotome formation despite the high spatiotemporal stochasticity of fusion events
Dev Cell. S1534-5807(22)00563-9. doi: 10.1016/j.devcel.2022.08.002.
[Link]
Larocque, G. & Royle, S.J. (2022)
Integrating intracellular nanovesicles into integrin trafficking pathways and beyond
Cell. Mol. Life Sci. 79: 335. doi: doi.org/10.1007/s00018-022-04371-6
[Link]
Koester, D.V., Bhat, A., Talluri, S., & Mayor S. (2022)
Reconstitution of Membrane-tethered Minimal Actin Cortices on Supported Lipid Bilayers
J Vis Exp. 12;(185). doi: 10.3791/63968.
[Link]
Hatano T, Palani S, Papatziamou D, Salzer R, Souza DP, Tamarit D, Makwana M, Potter A, Haig A, Xu W, Townsend D, Rochester D, Bellini D, Hussain HMA, Ettema TJG, Lowe J, Baum B, Robinson NP, Balasubramanian M. (2022)
Asgard archaea shed light on the evolutionary origins of the eukaryotic ubiquitin-ESCRT machinery
Nat Commun.13(1):3398. doi: 10.1038/s41467-022-30656-2
[Link]
Ferrandiz, N., Downie, L., Starling, G.P. and Royle, S.J. (2022)
Endomembranes promote chromosome missegregation by ensheathing misaligned chromosomes
J Cell Biol 2022 Jun 6;221(6). pii:jcb.202203021. doi: 10.1083/jcb.202203021
[Link]
Alvarez, Y. & Smutny, M. (2022)
Emerging Role of Mechanical Forces in Cell Fate Acquisition
Front Cell Dev Biol. 10:864522. doi: 10.3389/fcell.2022.864522
[Link]
Harrison JU, Sen O, McAinsh AD, Burroughs NJ. (2022)
Kinetochore tracking in 3D from lattice light sheet imaging data with KiT.
Bioinformatics. 2022 May 17:btac330. doi: 10.1093/bioinformatics/btac330.
[Link]
Wontakal SN, Britto M, Zhang H, Han Y, Gao C, Tannenbaum S, Durham BH, Lee
MT, An X, Mishima M. (2022)
RACGAP1 variants in a sporadic case of CDA III implicate the dysfunction of centralspindlin as the basis of the disease.
Blood. 2022 Mar 3;139(9):1413-1418. doi: 10.1182/blood.2021012334.
[Link]
Mahabaleshwar H, Asharani PV, Loo TY, Koh SY, Pitman MR, Kwok S, Ma J, Hu B, Lin F, Li Lok X, Pitson SM, Saunders TE, Carney TJ. (2022)
Slit-Robo signalling establishes a Sphingosine-1-phosphate gradient to polarise fin mesenchyme
EMBO Rep. 23(8):e54464. doi: 10.15252/embr.202154464.
[Link]
Lai JKH, Toh PJY, Cognart HA, Chouhan G, Saunders TE. (2022)
DNA-damage induced cell death in yap1;wwtr1 mutant epidermal basal cells
Elife. 11:e72302. doi: 10.7554/eLife.72302.
[Link]
Karkali K, Tiwari P, Singh A, Tlili S, Jorba I, Navajas D, Muñoz JJ, Saunders TE, Martin-Blanco E. (2022)
Condensation of the Drosophila nerve cord is oscillatory and depends on coordinated mechanical interactions
Dev Cell. 57(7):867-882.e5. doi: 10.1016/j.devcel.2022.03.007.
[Link]
de-Carvalho J, Tlili S, Hufnagel L, Saunders TE, Telley IA. (2022)
Aster repulsion drives short-ranged ordering in the Drosophila syncytial blastoderm
Development. 149(2):dev199997. doi: 10.1242/dev.199997.
[Link]
Germanova TE, Roscioli E, Harrison JU, McAinsh AD, Burroughs NJ. (2021)
Subcellular Euclidean distance measurements with multicolor fluorescence localization
imaging in cultured cells.
STAR Protoc. 2021 Nov 15;2(4):100774. doi: 10.1016/j.xpro.2021.100774.
[Link]
Blackley DG, Cooper JH, Pokorska P, Ratheesh A. (2021)
Mechanics of developmental migration.
Semin Cell Dev Biol. 2021 Dec;120:66-74. doi: 10.1016/j.semcdb.2021.07.002.
[Link]
Inman A, Smutny M. (2021)
Feeling the force: Multiscale force sensing and transduction at the cell-cell interface.
Semin Cell Dev Biol. 2021 Dec;120:53-65. doi: 10.1016/j.semcdb.2021.06.006.
[Link]
Sen O, Harrison JU, Burroughs NJ, McAinsh AD. (2021)
Kinetochore life histories reveal an Aurora-B-dependent error correction mechanism in anaphase.
Dev Cell. 2021 Nov 22;56(22):3082-3099.e5. doi: 10.1016/j.devcel.2021.10.007.
[Link]
Baker, A.N.; Richards, S.-J.; Pandey, S.; Guy, C.; Ahmad, A.; Hasan, M.; Biggs, C.; Georgiou, P.; Zwetsloot, A.; Straube, A.; Dedola, S.; Field, R.; Grammatopoulos, D.; Anderson, N.; Walker, M.; and Gibson, M. (2021)
A Glycan-based Flow-Through Device for the Detection of SARS-COV-2.
ACS Sensors 2021, doi: 10.1021/acssensors.1c01470
[Link]
Smith, S.M., Larocque, G., Wood, K.M., Morris, K.L., Roseman, A.M., Sessions, R.B., Royle, S.J. & Smith, C.J.
(2021)
Multi-modal adaptor-clathrin contacts drive coated vesicle assembly.
EMBO J e108795.
doi: 10.15252/embj.2021108795
[Link]
Yin Ho Vong, Lavanya Sivashanmugam, Rebecca Leech, Andreas Zaucker, Alex Jones, Karuna Sampath
(2021)
The RNA-binding protein Igf2bp3 is critical for embryonic and germline development in zebrafish
.
PLoS Genet 17(7): e1009667.
doi: 10.1371/journal.pgen.1009667
[Link]
Larocque, G., Moore, D.J., Sittewelle, M., Kuey, C., Hetmanski, J.H.R, La-Borde, P.J., Wilson, B.J., Clarke, N.I., Caswell, P.T. and Royle, S.J
(2021)
Intracellular nanovesicles mediate α5β1 integrin trafficking during cell migration.
J Cell Biol 220 (10): e202009028.
doi: 10.1083/jcb.202009028
[Link]
Palani S, Ghosh S, Ivorra-Molla E, Clarke S, Suchenko A, Balasubramanian MK, Koester DV.
(2021)
Calponin-homology domain mediated bending of membrane associated actin filaments.
eLife 10:e61078.
doi: 10.7554/eLife.61078
[Link]
Gharanei S, Fishwick K, Peter Durairaj R, Jin T, Siamantouras E, Liu K-K, Straube A, Lucas ES, Weston CJ, Rantakari P, Salmi M, Jalkanen S, Brosens JJ and Tan BK
(2021)
Vascular Adhesion Protein-1 Determines the Cellular Properties of Endometrial Pericytes.
Front. Cell Dev. Biol. 8:621016.
doi: 10.3389/fcell.2020.621016
[Link]
Malek, S., Köster, D.V. (2021)
The Role of Cell Adhesion and Cytoskeleton Dynamics in the Pathogenesis of the Ehlers-Danlos Syndromes and Hypermobility Spectrum Disorders
Frontiers in Cell and Developmental Biology
doi: 10.3389/fcell.2021.649082
[Link]
M. del Mar Aguilo-Ferretjans, R. Bosch, R. J. Puxty, M. Latva, V. Zadjelovic, A. Chhun, D. Sousoni, M. Polin, D. J. Scanlan, and J. A. Christie-Oleza (2021)
Pili allow dominant marine cyanobacteria to avoid sinking and evade predation.
Nature Communications 12, 1857
doi: https://doi.org/10.1038/s41467-021-22152-w
[Link]
Maruyama Y, Sugawa M, Yamaguchi S, Davies T, Osaki T, Kobayashi T, Yamagishi M, Takeuchi S, Mishima M, Yajima J.
M. del Mar Aguilo-Ferretjans, R. Bosch, R. J. Puxty, M. Latva, V. Zadjelovic, A. Chhun, D. Sousoni, M. Polin, D. J. Scanlan, and J. A. Christie-Oleza (2021)
CYK4 relaxes the bias in the off-axis motion by MKLP1 kinesin-6.
Commun Biol. 4:180. doi: 10.1038/s42003-021-01704-2
[Link]
I. Lopez-Grobas, M. Polin*, and M. Asally* (2021)
Swarming bacteria undergo localized dynamic phase transition to form stress-induced biofilms.
(* Joint last authors). eLife 10, e62632 doi: 10.7554/eLife.62632
[Link]
See also eLife‘s Press Release
H. L. Devereux, C. R. Twomey, M. S. Turner and S. Thutupalli (2021)
Whirligig beetles as corralled active Brownian particles
J. R. Soc. Interface, 18, 20210114 doi: 10.1098/rsif.2021.0114
[Link]
A. E. B. T. King and M.S.Turner (2021)
Non-local interactions in collective motion,
R. Soc. Open Sci. 8, 201536
[Link]
Ryan*, E.L., Shelford*, J., Massam-Wu, T., Bayliss, R. & Royle, S.J. (2020)
Defining endogenous TACC3–chTOG–clathrin–GTSE1 interactions at the mitotic spindle using induced relocalization
Journal of Cell Science 134(3):jcs255794 [Link]
Algirdas Toleikis, Nicholas J. Carter & Robert A. Cross (2020)
Backstepping mechanism of kinesin
Biophysical Journal 119, 1984-1994
doi.org/10.1016/j.bpj.2020.09.034
[Link]
Roscioli E, Germanova TE, Smith CA, Embacher PA, Erent M, Thompson AI, Burroughs NJ, McAinsh AD. (2020)
Ensemble-Level Organization of Human Kinetochores and Evidence for Distinct Tension and Attachment Sensors
Cell Reports 31(4):107535
doi.10.1016/j.celrep.2020.107535
[Link]
Yean Ming Chew & Robert A. Cross (2020)
Molecular Motors: Kif14s disordered dongle
Current Biology 30 R988-R990
DOI: 10.1016/j.cub.2020.06.095
[Link]
Piermarco Fonda, Sami C Al-Izzi, Luca Giomi, Matthew S Turner (2020)
Measuring Gaussian Rigidity Using Curved Substrates.
Phys Rev Lett 125(18):188002. doi: 10.1103/PhysRevLett.125.188002.
[Link]
Alexander N. Baker, Sarah-Jane Richards, Collette S. Guy, Thomas R. Congdon, Muhammad Hasan, Alexander J. Zwetsloot, Angelo Gallo, Józef R. Lewandowski, Phillip J. Stansfeld, Anne Straube, Marc Walker, Simona Chessa, Giulia Pergolizzi, Simone Dedola, Robert A. Field, and Matthew I. Gibson (2020)
The SARS-COV-2 Spike Protein Binds Sialic Acids and Enables Rapid Detection in a Lateral Flow Point of Care Diagnostic Device.
ACS Central Science 6 (11): 2046–2052 doi: 10.1021/acscentsci.0c00855
[Link]
Al-Izzi, S.C. Sens, P., Turner, M.S. and Komura, S. (2020)
Dynamics of passive and active membrane tubes
Soft Matter 2020 doi: 10.1039/d0sm01290d
[Link]
Francesca Bottanelli, Bruno Cadot, Felix Campelo, Scott Curran, Patricia M. Davidson, Gautam Dey, Ishier Raote, Anne Straube and Matthew P. Swaffer (2020)
Science during lockdown – from virtual seminars to sustainable online communities
Journal of Cell Science 2020 133: jcs249607
[Link]
Nida Siddiqui and Anne Straube (2020)
The Kinesin-3 Family: Long-Distance Transporters
in: The Kinesin Superfamily Handbook: Transporter, Creator, Destroyer, ed. Claire T. Friel, CRC Press, 2020
[Link to Book] [Open Access Chapter]
Auckland, P., Roscioli, E., Coker, H.L.E., McAinsh, A.D. (2020)
CENP-F stabilizes kinetochore-microtubule attachments and limits dynein stripping of corona cargoes.
Journal of Cell Biology. Vol. 219 No. 5 e201905018
[Link]
Mosby, L.S., Hundt, N., Young, G., Fineberg, A., Polin, M., Mayor, S., Kukura, P. & Köster, D.V. (2020)
Myosin II filament dynamics in actin networks revealed with interferometric scattering microscopy.
Biophysical Journal. 2020.02.025. doi: 10.1016/j.bpj.2020.02.025.
[Link]
Palani S., Köster D., Balasubramanian M., (2020)
Phosphoregulation of tropomyosin-actin interaction revealed using a genetic code expansion strategy.
Wellcome Open Research. doi: 10.12688/wellcomeopenres.16082.1.
[Link]
Mosby L., Polin M., Köster D. (2020)
A Python based automated tracking routine for myosin II filaments.
Journal of Physics D: Applied Physics. doi: 10.1088/1361-6463/ab87bf.
[Link]
Das, A.*, Bhat, A.*, Sknepnek, R., Köster, D., Mayor, S., Rao, M (2020)
Stratification relieves constraints from steric hindrance in the generation of compact actomyosin asters at the membrane cortex.
Science Advances. doi: 10.1126/sciadv.aay6093.
[Link]
Koester, D.V. (2020)
Pulling of Tethers from the Cell Plasma Membrane Using Optical Tweezers.
Methods Mol Biol. 2169:167-174. doi: 10.1007/978-1-0716-0732-9_15.
[Link]
Chapa-Y-Lazo, B., Hamanaka, M., Wray, A., Balasubramanian, M.K., & Mishima, M. (2020)
Polar relaxation by dynein-mediated removal of cortical myosin II.
J Cell Biol. 219(8). pii: e201903080. doi: 10.1083/jcb.201903080.
[Link]
Rushworth, L.K., Hewit, K., Munnings-Tomes, S., Somani, S., James, D., Shanks, E., Dufes, C., Straube, A., Patel, R., & Leung, H.Y. (2020)
Repurposing screen identifies mebendazole as a clinical candidate to synergise with docetaxel for prostate cancer treatment.
Br J Cancer. 122(4):517-527. doi: 10.1038/s41416-019-0681-5.
[Link]
Larocque, G., La-Borde, P.J., Clarke, N.I., Carter, N.J. & Royle, S.J. (2020)
Tumor Protein D54 defines a new class of intracellular transport vesicles.
The Journal of Cell Biology. 219: e201812044. doi: 10.1083/jcb.201812044.
[Link]
Jeanneret, R., Pushkin, D.O. & Polin, M. (2019)
Confinement Enhances the Diversity of Microbial Flow Fields..
Phys Rev Lett. 123(24):248102. doi: 10.1103/PhysRevLett.123.248102.
[Link]
Küey, C., Larocque, G., Clarke, N.I & Royle, S.J. (2019)
Unintended inhibition of protein function using GFP nanobodies in human cells.
Journal of Cell Science. 132: jcs234955. doi: 10.1242/jcs.234955.
[Link]
Capalbo, L., Bassi, Z.I., Geymonat, M., Todesca, S., Copoiu, L., Enright, A.J., Callaini, G., Riparbelli, M.G., Yu, L., Choudhary, J.S., Ferrero, E., Wheatley, S., Douglas, M.E., Mishima, M. & D Avino, P.P. (2019)
The midbody interactome reveals unexpected roles for PP1 phosphatases in cytokinesis.
Nat Commun. 10(1):4513. doi: 10.1038/s41467-019-12507-9.
[Link]
Adib R, Montgomery JM, Atherton J, O Regan L, Richards MW, Straatman KR, Roth
D, Straube A, Bayliss R, Moores CA, Fry AM. (2019)
Mitotic phosphorylation by NEK6 and NEK7 reduces the microtubule affinity of EML4 to promote chromosome congression.
Sci Signal. 12(594):eaaw2939. doi: 10.1126/scisignal.aaw2939.
[Link]
Shah, P., Chaumet, A., Royle, S.J. & Bard, F.A. (2019)
The NAE Pathway: Autobahn to the Nucleus for Cell Surface Receptors.
Cells, 33: 119-20. doi: 10.3390/cells8080915.
[Link]
Clare Garcin and Anne Straube (2019)
Microtubules in cell migration
Essays in Biochemistry 63(5): 509-520.
doi: 10.1042/EBC20190016
[Link]
N. Siddiqui*, A.J. Zwetsloot*, A. Bachmann, D. Roth, H. Hussain, J. Brandt, I. Kaverina & A. Straube (2019)
PTPN21 and Hook3 relieve KIF1C autoinhibition and activate intracellular transport
Nature Communications, Volume 10, Article number: 2693 doi: 10.1038/s41467-019-10644-9
[Link]
O. Von Loeffelholz, A. Peña, D.R. Drummond, R.A. Cross, C.A. Moores (2019)
4.5 Å Cryo-EM Structure of Yeast Kinesin-5-Microtubule Complex Reveals a Distinct Binding Footprint and Mechanism of Drug Resistance
J. Mol. Biol. doi: 10.1016/j.jmb.2019.01.011
[Link]
Bazarova, A., Nieduszynski, C.A., Akerman, I. and Burroughs, N.J. (2019)
Bayesian inference of origin firing time distributions, origin interference and licencing probabilities from Next Generation Sequencing data
Nucleic Acids Research. 2019, 47:2229–2243 [Link]
Cheffings, T.H. Burroughs, N.J. and Balasubramanian, M.K. (2019)
Actin turnover ensures uniform tension distribution during cytokinetic actomyosin ring contraction
Mol. Biol. Cell. 2019, 30(8):933-941. [Link]
Stanfield, Z., Lai, P.F., Lei, K., Johnson, M.R., Blanks, A.M., Romero, R., Chance, M.R., Mesiano, S. and Koyutürk, M. (2019)
Myometrial Transcriptional Signatures of Human Parturition
Front. Genet. [Link]
Syed, J., Palani, S., Clarke, S.T., Asad, Z., Bottrill, A.R., Jones, A.M.E., Sampath, K. and Balasubramanian, M.K. (2019)
Expanding the Zebrafish Genetic Code through Site-Specific Introduction of Azido-lysine, Bicyclononyne-lysine, and Diazirine-lysine
Int. J. Mol. Sci. [Link]
Zaucker, A, Kumari, P and K Sampath (2019)
Zebrafish embryogenesis - A framework to study regulatory RNA elements in development and disease
Developmental Biology [Link]
Dewulf, M., Köster, D., Sinha, B., Viaris de Lesegno, C., Chambon, V., Bigot, A., Bensalah, M., Negroni, E., Tardif, N., Podkalicka, J., Johannes, L., Nassoy, P., Butler-Browne, G., Lamaze, C. and Blouin, C.M. (2019)
Dystrophy-associated caveolin-3 mutations reveal that caveolae couple IL6/STAT3 signaling with mechanosensing in human muscle cells.
Nature Communications, 10 (1) 1974 [Link]
Ditlev, J.A., Vega, A.R., Köster, D.V., Su, X., Tani, T., Lakoduk, A.M., Vale, R.D., Mayor, S., Jaqaman, K., and Rosen, M.K. (2019)
A composition-dependent molecular clutch between T cell signaling condensates and actin
eLife 8:e42695
[Link]
Cross, R.A. (2019)
Microtubule lattice plasticity
Current Opinion in Cell Biology, doi: 10.1242/jcs.219550
[Link]
S Palani, DV Köster, T Hatano, A Kamnev, T Kanamaru, HR Brooker, ... (2019)
Phosphoregulation of tropomyosin is crucial for actin cable turnover and division site placement
Journal of Cell Biology 218 (11), 3548-3559
J Syed, S Palani, ST Clarke, Z Asad, AR Bottrill, AME Jones, K Sampath, ... (2019)
Expanding the zebrafish genetic code through site-specific introduction of azido-lysine, bicyclononyne-lysine, and diazirine-lysine
International journal of molecular sciences 20 (10), 2577
TH Cheffings, NJ Burroughs, MK Balasubramanian (2019)
Actin turnover ensures uniform tension distribution during cytokinetic actomyosin ring contraction
Molecular biology of the cell 30 (8), 933-941
JR Dunford, EJ Lutton, J Atia, AM Blanks, HA van den Berg (2019)
Computational physiology of uterine smooth muscle
Science progress 102 (2), 103-126
Z Stanfield, PF Lai, K Lei, MR Johnson, AM Blanks, R Romero, ... (2019)
Corrigendum: Myometrial Transcriptional Signatures of Human Parturition
Frontiers in Genetics 10, 515
Z Stanfield, PF Lai, K Lei, MR Johnson, AM Blanks, R Romero, ... (2019)
Myometrial transcriptional signatures of human parturition
Frontiers in genetics 10, 185
Jurić I, Hibberd JM, Blatt M, Burroughs NJ. (2019)
Computational modelling predicts substantial carbon assimilation gains for C3 plants with a single-celled C4 biochemical pump.
PLoS Comput Biol. 2019 Sep 30;15(9):e1007373. doi: 10.1371/journal.pcbi.1007373. eCollection 2019 Sep. PMID: 31568503 Free PMC article.
Bazarova A, Nieduszynski CA, Akerman I, Burroughs NJ. (2019)
Bayesian inference of origin firing time distributions, origin interference and licencing probabilities from Next Generation Sequencing data.
Nucleic Acids Res. 2019 Mar 18;47(5):2229-2243. doi: 10.1093/nar/gkz094. PMID: 30859196 Free PMC article.
AJ Pardal, F Fernandes-Duarte, AJ Bowman (2019)
The histone chaperoning pathway: from ribosome to nucleosome
Essays in Biochemistry 63 (1), 29-43
P Baniukiewicz, EJ Lutton, S Collier, T Bretschneider (2019)
Generative Adversarial Networks for Augmenting Training Data of Microscopic Cell Images
Frontiers in Computer Science 1, 10
O von Loeffelholz, A. Peña, DR Drummond, R Cross, CA Moores (2019)
Cryo-EM structure (4.5-Å) of yeast kinesin-5–microtubule complex reveals a distinct binding footprint and mechanism of drug resistance
Journal of molecular biology 431 (4), 864-872
C Halliday, K Billington, Z Wang, R Madden, S Dean, JD Sunter, ... (2019)
Cellular landmarks of Trypanosoma brucei and Leishmania mexicana
Molecular and biochemical parasitology 230, 24-36
S Dean, F Moreira-Leite, K Gull (2019)
Basalin is an evolutionarily unconstrained protein revealed via a conserved role in flagellum basal plate function
Elife 8, e42282
D Sage, TA Pham, H Babcock, T Lukes, T Pengo, J Chao, R Velmurugan, ... (2019)
Super-resolution fight club: assessment of 2D and 3D single-molecule localization microscopy software
Nature methods 16 (5), 387-395
AJ Perez, Y Cesbron, SL Shaw, J Bazan Villicana, HCT Tsui, MJ Boersma, ... (2019)
Movement dynamics of divisome proteins and PBP2x:FtsW in cells of Streptococcus pneumoniae
Proceedings of the National Academy of Sciences 116 (8), 3211-3220
N Stoop, N Waisbord, V Kantsler, V Heinonen, JS Guasto, J Dunkel (2019)
Disorder-induced topological transition in porous media flow networks
Journal of Non-Newtonian Fluid Mechanics 268, 66-74
R Mok, J Dunkel, V Kantsler (2019)
Geometric control of bacterial surface accumulation
Physical Review E 99 (5), 052607
S Kim, JM Kalappurakkal, S Mayor, MK Rosen (2019)
Phosphorylation of nephrin induces phase separated domains that move through actomyosin contraction
Molecular Biology of the Cell 30 (24), 2996-3012
JJ Thottacherry, M Sathe, C Prabhakara, S Mayor (2019)
Spoiled for choice: diverse endocytic pathways function at the cell surface
Annual review of cell and developmental biology 35 (1), 55-84
JA Ditlev, AR Vega, DV Köster, X Su, T Tani, AM Lakoduk, RD Vale, ... (2019)
A composition-dependent molecular clutch between T cell signaling condensates and actin
Elife 8, e42695
N Mateos Estevez, P Sil, C Manzo, J Torreno-Pina, S Mayor, ... (2019)
High density single particle tracking reveals nano-and meso-scale dynamic organization of plasma membrane receptors in living cells
EUROPEAN BIOPHYSICS JOURNAL WITH BIOPHYSICS LETTERS 48, S153-S153
S Krishnamoorthy, S Mayor, A Prabhakar (2019)
Mode re-locking in an RF detuned actively mode-locked fiber ring laser
2019 Conference on Lasers and Electro-Optics Europe & European Quantum …
M Kumar, S Sharma, P Sil, M Kushwaha, S Mayor, RA Vishwakarma, ... (2019)
C–H Arylation of N‐Heteroarenes under Metal‐Free Conditions and its Application towards the Synthesis of Pentabromo‐ and Pentachloropseudilins
European Journal of Organic Chemistry 2019 (22), 3591-3598
JM Kalappurakkal, AA Anilkumar, C Patra, TS van Zanten, MP Sheetz, ... (2019)
Integrin mechano-chemical signaling generates plasma membrane nanodomains that promote cell spreading
Cell 177 (7), 1738-1756. e23
A Hemalatha, S Mayor (2019)
Recent advances in clathrin-independent endocytosis
F1000Research 8, F1000 Faculty Rev-138
K Baker, IA Gyamfi, GI Mashanov, JE Molloy, MA Geeves, DP Mulvihill (2019)
TORC2-Gad8-dependent myosin phosphorylation modulates regulation by calcium
elife 8, e51150
S Khan, KH Downing, JE Molloy (2019)
Architectural dynamics of CaMKII-actin networks
Biophysical journal 116 (1), 104-119
K Valoskova, J Biebl, M Roblek, S Emtenani, A Gyoergy, M Misova, ... (2019)
A conserved major facilitator superfamily member orchestrates a subset of O-glycosylation to aid macrophage tissue invasion
Elife 8, e41801
C Küey, G Larocque, NI Clarke, SJ Royle (2019)
Unintended perturbation of protein function using GFP nanobodies in human cells
Journal of cell science 132 (21), jcs234955
G Larocque, PJ La-Borde, NI Clarke, NJ Carter, SJ Royle< (2019)br>
Tumor protein D54 defines a new class of intracellular transport vesicles
Journal of Cell Biology 219 (1), e201812044
J Syed, S Palani, ST Clarke, Z Asad, AR Bottrill, AME Jones, K Sampath, .. (2019)
Expanding the zebrafish genetic code through site-specific introduction of azido-lysine, bicyclononyne-lysine, and diazirine-lysine
International journal of molecular sciences 20 (10), 2577
X Yi, J Yu, C Ma, G Dong, W Shi, H Li, L Li, L Luo, K Sampath, H Ruan, ... (2019)
The effector of Hippo signaling, Taz, is required for formation of the micropyle and fertilization in zebrafish
PLoS Genetics 15 (1), e1007408
S Tlili, J Yin, JF Rupprecht, MA Mendieta-Serrano, G Weissbart, N Verma, ... (2019)
Shaping the zebrafish myotome by intertissue friction and active stress
Proceedings of the National Academy of Sciences 116 (51), 25430-25439
TE Saunders, PW Ingham (2019)
Open questions: how to get developmental biology into shape?
BMC biology 17, 1-3
O Hamant, T Saunders, V Viasnoff (2019)
Celebrate sustainable travel at conferences
Nature 573 (7774), 451-452
O Hamant, T Saunders, V Viasnoff (2019)
Seven steps to make travel to scientific conferences more sustainable
Nature 573 (7774), 451-453
H Connahs, S Tlili, T Saunders, A Monteiro (2019)
The people behind the papers-Heidi Connahs, Sham Tlili, Timothy Saunders and Antonia Monteiro
DEVELOPMENT 146 (9)
H Connahs, S Tlili, J van Creij, TYJ Loo, TD Banerjee, TE Saunders, ... (2019)
Activation of butterfly eyespots by Distal-less is consistent with a reaction-diffusion process
Development 146 (9), dev169367
O Hamant, T Saunders, V Viasnoff (2019)
CAREERS Celebrate sustainable
H Connahs, S Tlili, J van Creij, TYJ Loo, TD Banerjee, TE Saunders, ... (2019)
Distal-less activates butterfly eyespots consistent with a reaction diffusion process
Development 146, dev169367
D Čapek, M Smutny, AM Tichy, M Morri, H Janovjak, CP Heisenberg (2019)
Light-activated Frizzled7 reveals a permissive role of non-canonical wnt signaling in mesendoderm cell migration
Elife 8, e42093
C Garcin, A Straube (2019)
Microtubules in cell migration
Essays in biochemistry 63 (5), 509-520
R Adib, JM Montgomery, J Atherton, L O’Regan, MW Richards, ... (2019)
Mitotic phosphorylation by NEK6 and NEK7 reduces the microtubule affinity of EML4 to promote chromosome congression
Science signaling 12 (594), eaaw2939
N Siddiqui, AJ Zwetsloot, A Bachmann, D Roth, H Hussain, J Brandt, ... (2019)
PTPN21 and Hook3 relieve KIF1C autoinhibition and activate intracellular transport
Nature communications 10 (1), 2693
D Roth, BP Fitton, NP Chmel, N Wasiluk, A Straubev (2019)
Spatial positioning of EB family proteins at microtubule tips involves distinct nucleotide-dependent binding properties
Journal of Cell Science 132 (4), jcs219550
Charlesworth, Henry J., Turner, Matthew S. (2019)
Intrinsically motivated collective motion
Proceedings of the National Academy of Sciences of the United States of America National Academy of Sciences 116 31 pp. 15362-15367 10.1073/pnas.1822069116

Slator P.J. & Burroughs, N.J. (2018)
A Hidden Markov Model for Detecting Confinement in Single-Particle Tracking Trajectories
Biophys J. 2018 Nov 6; 115(9): 1741–1754
[Link]

Lutton EJ, Lammers WJEP, James S, van den Berg HA, Blanks AM (2018)
Identification of uterine pacemaker regions at the myometrial-placental interface in the rat
J Physiol. 2018 Jul;596(14):2841-2852. doi: 10.1113/JP275688
[Link]

Liu, Y., Claydon, R., Polin, M. and Brumley, D.R. (2018)
Transitions in synchronization states of model cilia through basal-connection coupling
Journal of The Royal Society Interface, doi: 10.1098/rsif.2018.0450
[Link]
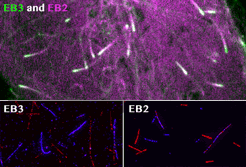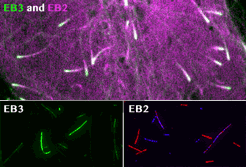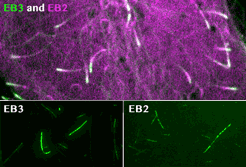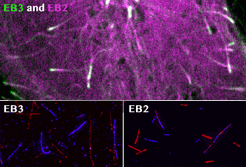
Roth, D., Fitton, B.P., Chmel, N., Wasiluk, N. and Straube, A. (2018)
Spatial positioning of EB family proteins at microtubule tips involves distinct nucleotide-dependent binding properties
Journal of Cell Science 132:jcs219550
doi: 10.1242/jcs.219550
[Link]
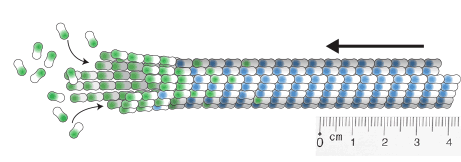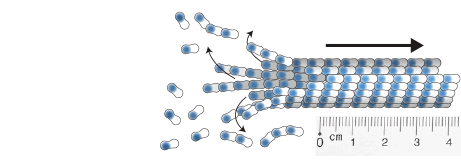
Zwetsloot, A.J., Tut, G. and Straube, A. (2018)
Measuring microtubule dynamics
Essays in Biochemistry 62(6):725-735, doi: 10.1042/EBC20180035
[Link]
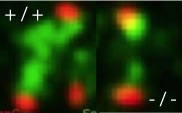
Currie CE, Mora-Santos M, Smith CA, McAinsh AD and Millar JBA. (2018)
Bub1 is not essential for the checkpoint response to unattached kinetochores in diploid human cells.
Current Biology 28, 929-930.
[Link]
John C Meadows, Liam J Messin, Anton Kamnev, Theresa C Lancaster, Mohan K Balasubramanian, Robert A Cross, Jonathan BA Millar (2018)
Opposing kinesin complexes queue at plus tips to ensure microtubule catastrophe at cell ends
EMBO reports (2018) e46196 doi:10.15252/embr.201846196
[Link]
Wood, L.A. & Royle, S.J. (2018)
Imaging "Hot-Wired" Clathrin-Mediated Endocytosis.
Methods Mol Biol. 1847: 83-94. doi: 10.1007/978-1-4939-8719-1_7.
[Link]

Clarke, N.I. & Royle, S.J. (2018)
Correlating light microscopy with serial block face scanning electron microscopy to study mitotic spindle architecture.
Methods Cell Biol. 145: 29-43. doi: 10.1016/bs.mcb.2018.03.010.
[Link]

Clarke, N.I. & Royle, S.J. (2018)
FerriTag is a new genetically-encoded inducible tag for correlative light-electron microscopy.
Nat Commun. 9: 2604. doi: 10.1038/s41467-018-04993-0.
[Link]
Baniukiewicz, P., Collier, S. & Bretschneider, T. (2018)
QuimP: analyzing transmembrane signalling in highly deformable cells.
Bioinformatics. bty169.
[Link]

Dudka, D., Noatynska, A., Smith, C.A, Liaudet, N., McAinsh, A.D. & Meraldi, P. (2018)
Complete microtubule–kinetochore occupancy favours the segregation of merotelic attachments.
Nat Commun. 9: 2042.
[Link]

McHugh, T., Drechsler, H., McAinsh, A.D., Carter, N.J. & Cross R.A. (2018)
Kif15 functions as an active mechanical ratchet.
Mol Biol Cell. mbcE18030151. doi: 10.1091/mbc.E18-03-0151.
[Link]
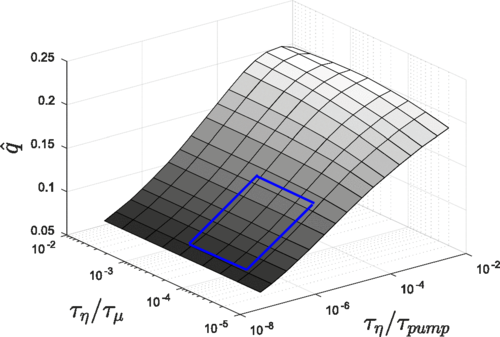
Al-Izzi, S.C., Rowlands, G., Sens, P & Turner, M.S. (2018)
Hydro-osmotic Instabilities in Active Membrane Tubes.
Phys Rev Lett. 120(13):138102. doi: 10.1103/PhysRevLett.120.138102.
[Link]

Peet, D.R., Burroughs, N.J. & Cross, R.A. (2018)
Kinesin expands and stabilizes the GDP-microtubule lattice.
Nat Nanotechnol doi: 10.1038/s41565-018-0084-4
[Link]
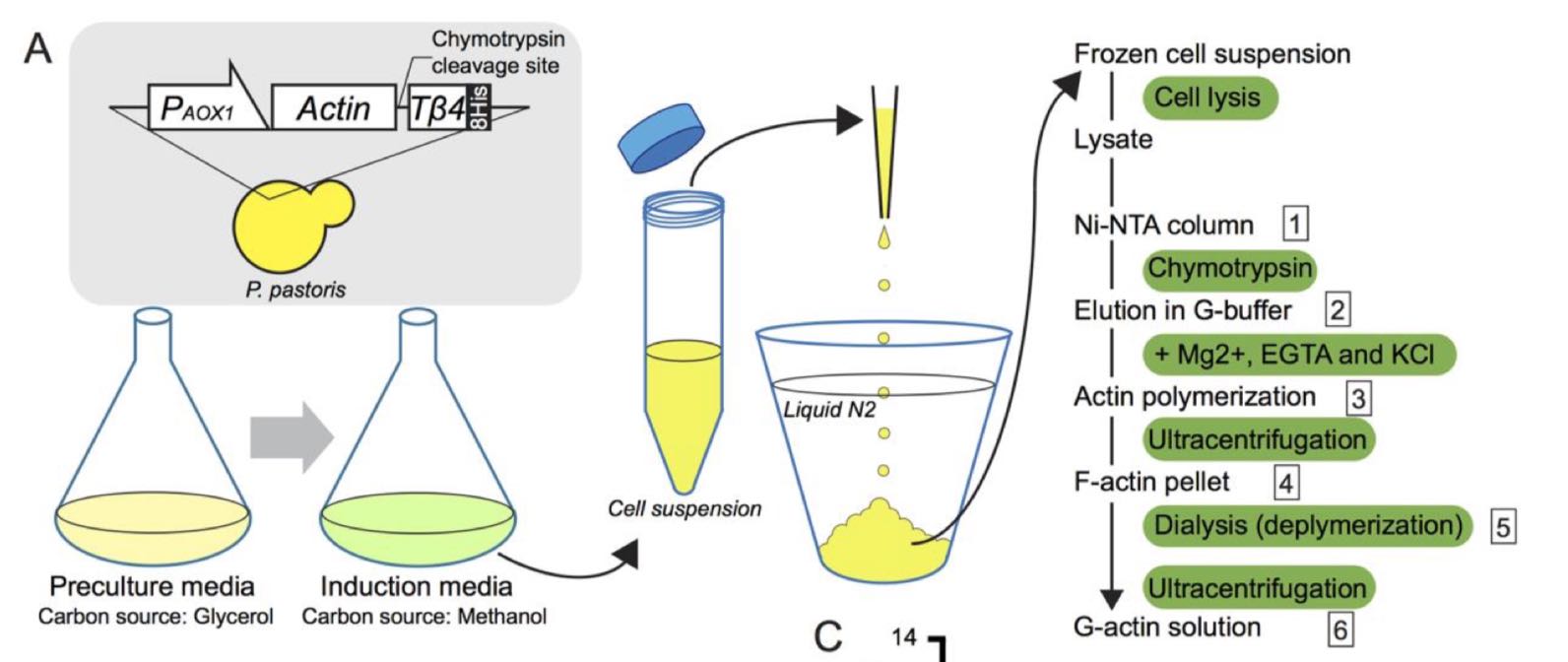
Hatano, T., Alioto, S., Roscioli, E., Palani, S., Clarke, S.T., Kamnev, A., Hernandez-Fernaud, J.R., Sivashanmugam, L., Chapa, Y.L.B., Jones, A.M.E., Robinson, R.C., Sampath, K., Mishima, M., McAinsh, A.D., Goode, B.L. & Balasubramanian, M.K. (2018)
Rapid production of pure recombinant actin isoforms in Pichia pastoris.
J Cell Sci doi: 10.1242/jcs.213827
[Link]
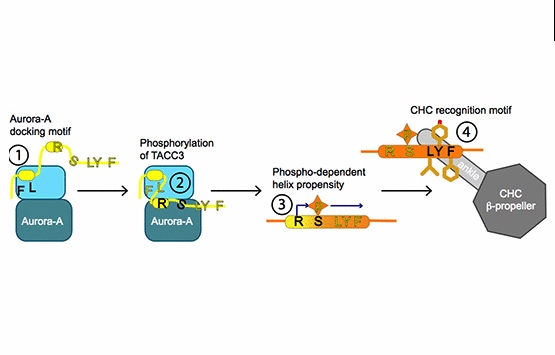
Burgess, S.G., Mukherjee, M., Sabir, S., Joseph, N., Gutierrez-Caballero, C., Richards, M.W., Huguenin-Dezot, N., Chin, J.W., Kennedy, E.J., Pfuhl, M., Royle, S.J., Gergely, F. & Bayliss, R. (2018)
Mitotic spindle association of TACC3 requires Aurora-A-dependent stabilization of a cryptic alpha-helix.
EMBO J doi:10.15252/embj.201797902
[Link]
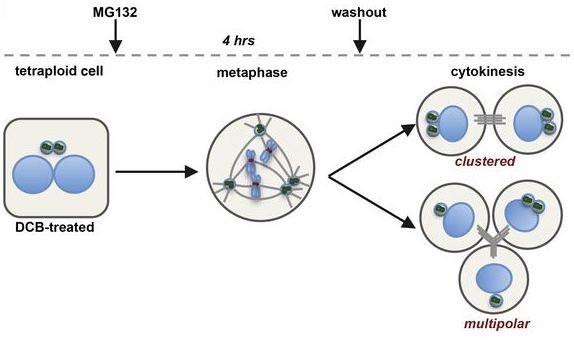
Rhys, A.D., Monteiro, P., Smith, C., Vaghela, M., Arnandis, T., Kato, T., Leitinger, B., Sahai, E., McAinsh, A., Charras, G. & Godinho, S.A. (2018)
Loss of E-cadherin provides tolerance to centrosome amplification in epithelial cancer cells.
J Cell Biol doi: 10.1083/jcb.201704102
[Link]
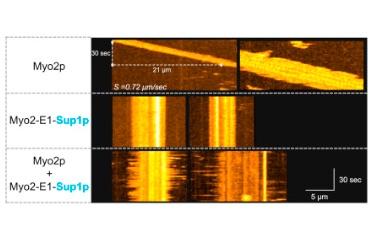
Palani, S., Srinivasan, R., Zambon, P., Kamnev, A., Gayathri, P. & Balasubramanian, M.K. (2018)
Steric hindrance in the upper 50 kDa domain of the motor Myo2p leads to cytokinesis defects in fission yeast.
J Cell Sci doi:10.1242/jcs.205625
[Link]

Mishima, M. (2017)
Preparation of centralspindlin as an active heterotetramer of kinesin and GAP subunits for in vitro structural and functional assays
Methods Cell Biol. 2017;137:371-385. doi: 10.1016/bs.mcb.2016.04.005
[Link]
 Rhys,
A.D., Monteiro, P., Smith, C. Vaghela, M., Arnandis,
T., Kato, T., Leitinger, B., Sahai, E., McAinsh, A.,
Charras, G. and Godinho, S.A. (2017)
Loss of E-cadherin provides tolerance to centrosome
amplification in epithelial cancer cells
Rhys,
A.D., Monteiro, P., Smith, C. Vaghela, M., Arnandis,
T., Kato, T., Leitinger, B., Sahai, E., McAinsh, A.,
Charras, G. and Godinho, S.A. (2017)
Loss of E-cadherin provides tolerance to centrosome
amplification in epithelial cancer cells
Journal of Cell Biology
[Link]
 Ottilie von Loeffelholz, Neil A. Venables, Douglas
R. Drummond, Miho Katsuki, Robert A. Cross & Carolyn A. Moores (2017)
Ottilie von Loeffelholz, Neil A. Venables, Douglas
R. Drummond, Miho Katsuki, Robert A. Cross & Carolyn A. Moores (2017)
Nucleotide– and Mal3-dependent changes in fission
yeast microtubules suggest a structural plasticity
view of dynamics
Nature Communications, 8: 2110
[Open]
 Meadows, J.C and Millar, J.B.A. (2017)
Meadows, J.C and Millar, J.B.A. (2017)
Some assembly required: Redefining the mitotic
checkpoint
Molecular & Cellular Oncology, 4(6):e1314238
[Link]
 Zaucker, A., Nagorska, A., Kumari, P., Hecker, N.,
Wang, Y., Huang, S., Cooper, L., Sivashanmugam, L.,
VijayKumar, S., Brosens, J., Gorodkin, J. and Sampath,
K. (2017)
Zaucker, A., Nagorska, A., Kumari, P., Hecker, N.,
Wang, Y., Huang, S., Cooper, L., Sivashanmugam, L.,
VijayKumar, S., Brosens, J., Gorodkin, J. and Sampath,
K. (2017)
Translational co-regulation of a ligand and
inhibitor by a conserved RNA element
Nucleic Acids Research, gkx938
[Link]
 Wood,
L.A., Larocque, G., Clarke, N.I., Sarkar, S. and
Royle, S.J. (2017)
Wood,
L.A., Larocque, G., Clarke, N.I., Sarkar, S. and
Royle, S.J. (2017)
New tools for “hot-wiring” clathrin-mediated
endocytosis with temporal and spatial precision
Journal of Cell Biology, 216: 4351-65; DOI:
10.1083/jcb.201702188
[Link]
 Sarkar, S., Ryan, E.L. and Royle, S.J. (2017)
Sarkar, S., Ryan, E.L. and Royle, S.J. (2017)
FGFR3–TACC3 cancer gene fusions cause mitotic
defects by removal of endogenous TACC3 from the
mitotic spindle
Open Biology, 7: 170080; DOI: 10.1098/rsob.170080
[Open]
 Siddiqui, N. and Straube, A. (2017)
Siddiqui, N. and Straube, A. (2017)
Intracellular cargo transport by kinesin-3 motors
Biochemistry (Moscow), Vol. 82, No. 7, pp. 803–815
Russian version: Biokhimiya, 2017, Vol. 82, No. 7, pp.
1047–1062
[open
access article]
 Collier, S., Paschke, P., Kay, R.R. and Bretschneider,
T. (2017)
Collier, S., Paschke, P., Kay, R.R. and Bretschneider,
T. (2017)
Image based modeling of bleb site selection
Scientific Reports, 7: 6692 doi:
10.1038/s41598-017-06875-9
[Open]
Chew, T.G., Huang, J., Palani, S., Sommese, R.,
Kamnev, A., Hatano, T., Gu, Y., Oliferenko, S.,
Sivaramakrishnan, S. and Balasubramanian, M.K. (2017)
Actin turnover maintains actin filament homeostasis
during cytokinetic ring contraction
Journal of Cell Biology doi: 10.1083/jcb.201701104
[Open]

Nixon*, F.M., Honnor*, T.R., Clarke, N.I., Starling,
G.P., Beckett, A.J., Johansen, A.M., Brettschneider,
J.A., Prior, I.A. and Royle, S.J. (2017)
Microtubule organization within mitotic spindles
revealed by serial block face scanning electron
microscopy and image analysis
Journal of Cell Science, 130: 1845-55. doi:
10.1242/jcs.203877
[Link]

Auckland, P., Clarke, N.I., Royle, S.J. and McAinsh,
A.D. (2017)
Congressing kinetochores progressively load Ska
complexes to prevent force-dependent detachment
Journal of Cell Biology. 216: 1623-39. doi:
10.1083/jcb.201607096.
[Link]
Lutton, E.J., Lammers, W.J., James, S., van den
Berg, H.A., Blanks, A.M. (2017)
A computational method for three-dimensional
reconstruction of the microarchitecture of
myometrial smooth muscle from histological sections
PLoS One. 12, e0173404 doi:
10.1371/journal.pone.0173404.
[Link]

Palani, S., Chew, T.G., Ramanujam, S., Kamnev, A.,
Harne, S., Chapa-Y-Lazo, B., Hogg, R., Sevugan, M.,
Mishra, M., Gayathri, P., and Balasubramanian, M.K.
(2017)
Motor Activity Dependent and Independent Functions
of Myosin II Contribute to Actomyosin Ring Assembly
and Contraction in Schizosaccharomyces pombe
Current Biology. doi: 10.1016/j.cub.2017.01.028.
[Link]
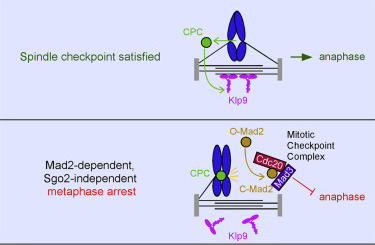
Meadows, J.C., Lancaster, T.C., Buttrick, G.J.,
Sochaj, A.M., Messin, L.J., Mora-Santos, M.D.,
Hardwick, K.G. and Millar, J.B.A. (2017)
Identification of a Sgo2-Dependent but
Mad2-Independent Pathway Controlling Anaphase Onset
in Fission Yeast
Cell Reports. 18(6):1422-1433. doi:
10.1016/j.celrep.2017.01.032.
[Link]
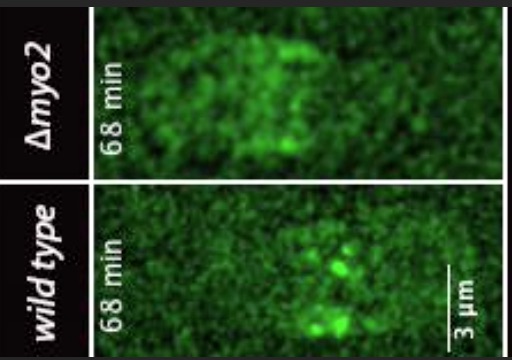
Zambon, P., Palani, S., Kamnev, A. and
Balasubramanian, M.K. (2017)
Myo2p is the major motor involved in actomyosin ring
contraction in fission yeast
Current Biology. 27(3):R99-R100. doi:
10.1016/j.cub.2016.12.024.
[Link]

Mishima, M. (2016)
Centralspindlin in Rappaport's cleavage signaling
Semin Cell Dev Biol. 2016 May;53:45-56. doi: 10.1016/j.semcdb.2016.03.006. Epub 2016 Mar 7. Review.
[Link]
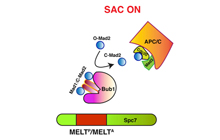
Mora-Santos MD, Hervas-Aguilar A, Sewart K,
Lancaster TC, Meadows JC, Millar JB (2016)
Bub3-Bub1 Binding to Spc7/KNL1 Toggles the Spindle
Checkpoint Switch by Licensing the Interaction of
Bub1 with Mad1-Mad2
Curr Biol. 2016 Oct 10;26(19):2642-2650. doi:
10.1016/j.cub.2016.07.040. PMID: 27618268
[Link]
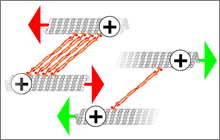
Mishan Britto, Adeline Goulet, Syeda Rizvi, Ottilie
von Loeffelholz, Carolyn A. Moores, and Robert A.
Cross (2016)
Schizosaccharomyces pombe kinesin-5 switches
direction using a steric blocking mechanism
PNAS DOI: 10.1073/pnas.1611581113
[Open]
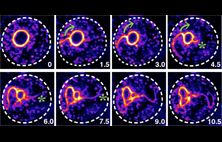
Junqi Huang, Ting Gang Chew, Ying Gu, Saravanan
Palani, Anton Kamnev, Douglas S. Martin, Nicholas J.
Carter, Robert A. Cross, Snezhana Oliferenko and Mohan
K Balasubramanian (2016)
Curvature-induced expulsion of actomyosin bundles
during cytokinetic ring contraction
eLife 2016;10.7554/eLife.21383
[Open]

Chris A. Smith, Andrew D. McAinsh and Nigel J.
Burroughs (2016)
Human kinetochores are swivel joints that mediate
microtubule attachments
eLife 10.7554/eLife.16159
[Open]
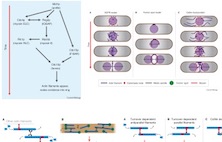
Thomas H. Cheffings, Nigel J. Burroughs and Mohan K.
Balasubramanian correspondence (2016)
Actomyosin Ring Formation and Tension Generation in
Eukaryotic Cytokinesis
Curr Biol 26: R719-R737
[Review]
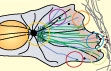
Ulrike Theisen and Anne Straube (2016)
Microtubules Regulate Cell Migration and Neuronal
Pathfinding
in: The Microtubule Cytoskeleton, Jens Lüders (ed),
pages 151-189 DOI: 10.1007/978-3-7091-1903-7_6
[Link]
Hervas-Aguilar A, Millar JB.
Mph1/MPS1 checks in at the kinetochore
Cell Cycle. 2016 May 18;15(10):1313-4. doi:
10.1080/15384101.2016.1159888. PMID: 27105354
[Link]
Robert A. Cross (2016)
Mechanochemistry of the kinesin-1 ATPase
Biopolymers DOI: 10.1002/bip.22862
[Open]
 Hauke
Drechsler and Andrew D. McAinsh (2016)
Hauke
Drechsler and Andrew D. McAinsh (2016)
Kinesin-12 motors cooperate to suppress microtubule
catastrophes and drive the formation of parallel
microtubule bundles
Proc Natl Acad Sci U S A. Mar 11
[Open
Access]
 Jonathan
W. Armond, Elina Vladimirou, Andrew D. McAinsh, and
Nigel J. Burroughs (2016)
Jonathan
W. Armond, Elina Vladimirou, Andrew D. McAinsh, and
Nigel J. Burroughs (2016)
KiT: A MATLAB package for kinetochore tracking
Bioinformatics First published online: February 15
[Open
Access]
 Frauke
Hussmann, Douglas R. Drummond, Daniel Peet, Douglas S.
Martin & Robert A. Cross (2016)
Frauke
Hussmann, Douglas R. Drummond, Daniel Peet, Douglas S.
Martin & Robert A. Cross (2016)
Alp7/TACC-Alp14/TOG generates long-lived,
fast-growing MTs by an unconventional mechanism
Nature Scientific Reports DOI: 10.1038/srep20653
[Open
Access]
 Joanna Andrecka, Jaime Ortega Arroyo, Katie Lewis,
Robert A. Cross, and Philipp Kukura (2016)
Joanna Andrecka, Jaime Ortega Arroyo, Katie Lewis,
Robert A. Cross, and Philipp Kukura (2016)
Label-free Imaging of Microtubules with Sub-nm
Precision Using Interferometric Scattering
Microscopy
Biophysical Journal 110 214-217
[Link]
 Vladimirou
E., Harry, E., Burroughs N. & McAinsh A.D. (2011)
Vladimirou
E., Harry, E., Burroughs N. & McAinsh A.D. (2011)
Springs, clutches and motors: driving forward
kinetochore mechanism by modelling
Chromosome Biology, 19 409-21
[link
to pdf] [pubmed
abstract]
*equal contribution **corresponding authors
 Amaro
AC, Samora CP, Holtackers R, Wang E, Kingston I,
Alonso M, Lampson L, McAinsh AD** and Meraldi P**
(2010)
Amaro
AC, Samora CP, Holtackers R, Wang E, Kingston I,
Alonso M, Lampson L, McAinsh AD** and Meraldi P**
(2010)
Molecular control of kinetochore-microtubule
dynamics and chromosome oscillation
Nature Cell Biology, 12 319-329
[link
to pdf] [pubmed
abstract]
 Jaqaman K*, King E*,
Amaro AC*, Winter JR*, Dorn JF, Elliott, HL,
Mchedlishvili N, McClelland SE, Porter IM, Posch M,
Toso A, Danuser G**, McAinsh AD**, Meraldi P**,
Swedlow JR** (2010)
Jaqaman K*, King E*,
Amaro AC*, Winter JR*, Dorn JF, Elliott, HL,
Mchedlishvili N, McClelland SE, Porter IM, Posch M,
Toso A, Danuser G**, McAinsh AD**, Meraldi P**,
Swedlow JR** (2010)
Kinetochore alignment within the metaphase plate is
regulated by centromere stiffness and microtubule
depolymerases
Journal of Cell Biology, 188 665-79
[link
to pdf] [pubmed
abstract]
*equal contribution **corresponding authors



















 Ottilie von Loeffelholz, Neil A. Venables, Douglas
R. Drummond, Miho Katsuki, Robert A. Cross & Carolyn A. Moores (2017)
Ottilie von Loeffelholz, Neil A. Venables, Douglas
R. Drummond, Miho Katsuki, Robert A. Cross & Carolyn A. Moores (2017)
 Meadows, J.C and Millar, J.B.A. (2017)
Meadows, J.C and Millar, J.B.A. (2017) Zaucker, A., Nagorska, A., Kumari, P., Hecker, N.,
Wang, Y., Huang, S., Cooper, L., Sivashanmugam, L.,
VijayKumar, S., Brosens, J., Gorodkin, J. and Sampath,
K. (2017)
Zaucker, A., Nagorska, A., Kumari, P., Hecker, N.,
Wang, Y., Huang, S., Cooper, L., Sivashanmugam, L.,
VijayKumar, S., Brosens, J., Gorodkin, J. and Sampath,
K. (2017) Sarkar, S., Ryan, E.L. and Royle, S.J. (2017)
Sarkar, S., Ryan, E.L. and Royle, S.J. (2017) Siddiqui, N. and Straube, A. (2017)
Siddiqui, N. and Straube, A. (2017) Collier, S., Paschke, P., Kay, R.R. and Bretschneider,
T. (2017)
Collier, S., Paschke, P., Kay, R.R. and Bretschneider,
T. (2017)













 Hauke
Drechsler and Andrew D. McAinsh (2016)
Hauke
Drechsler and Andrew D. McAinsh (2016) Jonathan
W. Armond, Elina Vladimirou, Andrew D. McAinsh, and
Nigel J. Burroughs (2016)
Jonathan
W. Armond, Elina Vladimirou, Andrew D. McAinsh, and
Nigel J. Burroughs (2016) Vladimirou
E., Harry, E., Burroughs N. & McAinsh A.D. (2011)
Vladimirou
E., Harry, E., Burroughs N. & McAinsh A.D. (2011) Amaro
AC, Samora CP, Holtackers R, Wang E, Kingston I,
Alonso M, Lampson L, McAinsh AD** and Meraldi P**
(2010)
Amaro
AC, Samora CP, Holtackers R, Wang E, Kingston I,
Alonso M, Lampson L, McAinsh AD** and Meraldi P**
(2010)